Have you ever thought about what you’d be like without a skeleton?
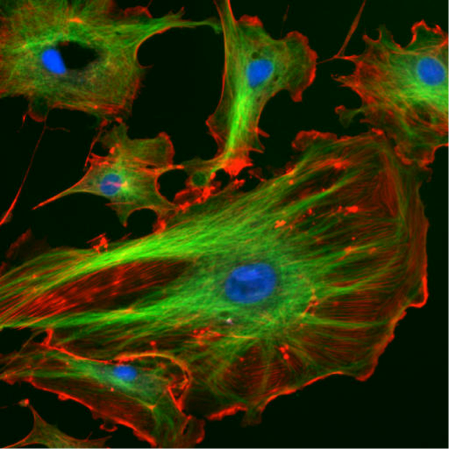
Our skeletons give us our structure and also allow us to move. Without the bones that make up our skeleton we’d just be a mass of tissues that couldn’t move. In much the same way, cells have something called a cytoskeleton to give them structure and direct their movement. Unlike with human skeletons, cells also use their cytoskeletons as an internal transportation system, shuttling material back and forth throughout the cell, and in cell division. To do these things, cells use three types of cytoskeletal elements: actin, microtubules, and intermediate filaments. Actin (red in Figure 1 below) is often found along the outside of the cell, whereas microtubules (green) can be seen throughout the cell.
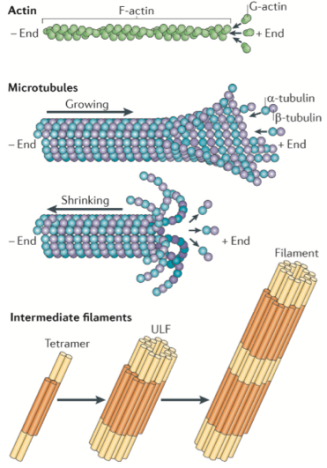
While the protein makeup of these three components is different (shown in Figure 2), what they have in common is that they are all long, thin structures. Because the cytoskeleton proteins are so diverse and so important, a lot of scientists want to study them. In addition, because the cytoskeleton is often involved in dynamic processes, researchers need special microscopy methods to use for imaging studies. One challenge is that the cytoskeletal components are so thin that they are below the diffraction limit of light, requiring the use of microscopy techniques that bring us close to or below that limit (see this previous post for an explanation of the diffraction limit of light).
There are many interactions beyond the cytoskeleton, such as protein-protein interactions, that scientists are interested in studying which involve features or species below the diffraction limit of light. This led to the development of several microscopy techniques called super-resolution fluorescence microscopy, which allow us to actually be able to see such tiny structures. One example is stochastic optical reconstruction microscopy (STORM) that researchers from the Center for Sustainable Nanotechnology used to study how nanoparticles bind to the surface of bacteria.2 There are other super-resolution microscopies (see examples in previous posts here and here), but one of the great benefits of using fluorescence microscopy is that it allows us to see only specific structures or interactions of interest within a cell by adding fluorescent molecules to only those components, which allows us to ignore everything else in the cell.
Given all the different kinds of super-resolution microscopy, how should scientists pick which option to use? In a review of the literature that I recently published with several coauthors,3 we compare different types of near-resolution and super-resolution fluorescence microscopy and discuss how each has been used to study the cytoskeleton. When deciding on a technique, it is important to consider the scientific question we want to investigate. First, because the biological processes involving the cytoskeleton are dynamic, we want to capture changes over time. However, it’s not always so simple: with the cytoskeleton we sometimes have to choose between seeing the interactions between structures very clearly (high spatial resolution) or seeing the interactions that are occurring very quickly (high temporal resolution). To differentiate between spatial and temporal resolution, imagine a cheetah. If we were interested in the spots on a cheetah we would want a high spatial resolution image to show all the spotty details. If we were interested in the mechanics of how they sprint, we would want a series of images taken in very fast succession. We probably couldn’t use the same camera to get both types of images.
Luckily, with different types of high-resolution fluorescence microscopy we can choose a technique best suited to the question we are trying to answer. If our questions are more related to cell structure, then scientists can work with fixed cells – cells that have been treated with certain chemicals to “freeze” them in their orientation at the time of interest. In that case, the researchers should choose high-resolution techniques that take longer to acquire images but do so with more detail. On the other hand, if the scientific questions are related to very quick interactions between parts of the cell then researchers should use techniques that gather data quickly but at the expense of fine detail.
Our review of imaging for the cytoskeleton covers two near-resolution fluorescence microscopy techniques (confocal and TIRF) and three super-resolution microscopy techniques (PALM, STORM, and STED).
- Confocal and TIRF (or total internal reflection fluorescence) microscopy give highly detailed images of specific regions of a cell. Using confocal microscopy, scientists can image “slices” of cells and it is suited to non-dynamic imaging of actin filaments (red in figure 1, and the thinnest strand in figure 2).
- Using TIRF microscopy, interactions that occur near the cell membrane can be imaged – it has been used to study the role of the cytoskeleton in cellular processes such as how an immune response works, what proteins or other molecules are being secreted from a cell, and how a cell moves.
- PALM and STORM function by taking a series of images where fluorescent probes are repeatedly “turned on and off” and then the image series is combined to make a very detailed image. You can see some cool images taken with STORM in Figure 3 below.
- On the other hand, STED uses a system of two lasers to create a small fluorescent area for targeted imaging.
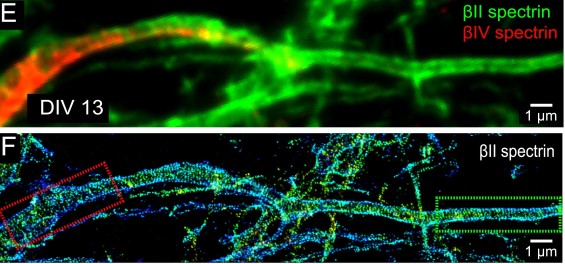
These super-resolution techniques allow scientists to gain a better understanding of how the cytoskeletal filaments themselves are assembled and regulated. The review that we wrote analyzes the pros and cons of each technique for various scientific questions, and we even make suggestions for experiments that scientists can do to fill knowledge gaps in the field! With the large toolset available, scientists will keep improving our understanding of the cytoskeleton and the many roles it plays in supporting cells.
REFERENCES:
- Mostowy, S. & Cossart, P. Septins: the fourth component of the cytoskeleton, Nature Reviews Molecular Cell Biology, 2012, 13, 183– 194. doi: 10.1038/nrm3284
- Jacobson, K., Gunsolus, I., Kuech, T.; Troiano, J., Melby, E., Lohse, S., Hu, D., Chrisler, W., Murphy, C., Orr, G., Geiger, F., Haynes, C., & Pedersen, J. Lipopolysaccharide density and structure governs the extent and distance of nanoparticle interaction with actual and model bacterial outer membranes. Environmental Science & Technology, 2015, 49 (17), 10642–10650 doi: 10.1021/acs.est.5b01841
- Finkenstaedt-Quinn, S., Qiu, T., Shin, K., & Haynes, C. Super-resolution imaging for monitoring cytoskeleton dynamics. Analyst, 2016, 141, 5674-5688. doi: 10.1039/C6AN00731G
- Zhong et al. Developmental mechanism of the periodic membrane skeleton in axons. eLife 2014;3:e04581. doi: 10.7554/eLife.04581
