Have bacteria ever made you sick?
If you answer no I’m not going to believe you. When most of us think about bacteria, we’re reminded of how miserable we were the last time we had strep throat or another bacterial illness. Of course there are many other things that can make people sick, but in a lot of situations bacteria are to blame. Because of that, we don’t often think of all the good that bacteria do for us, such as helping us digest food and absorb its nutrients. Bacteria also do a lot to maintain the environment.

Bacteria have many important environmental roles, such as converting nutrients in soil or water to a form that can be used by other organisms like plants. Because of this, bacteria are good indicators of the overall health of an environment – if the bacteria die off, there will be less available nutrients for higher organisms and they too will die. This is why scientists use bacteria in environmental studies. In the Center for Sustainable Nanotechnology, we’re interested in studying what impact nanomaterials that are used in everyday products may have when they’re released in the environment. For example, I recently published a paper on experiments done with eight other people where we investigated how a panel of diverse bacteria reacted when we exposed them to gold nanoparticles with a particular kind of wrapping called polyallylamine hydrochloride, or PAH (Fig. 2).1
We know that when nanoparticles interact with bacteria, they often stick to the bacterial surface and don’t actually enter the “body” of the bacterium. But just interacting with the surface is enough to cause toxicity. Previous work in the CSN has shown that on a class of bacteria known as Gram-negative bacteria, one type of molecule called lipopolysaccharides (LPS) helps nanoparticles stick to the bacterial surface kind of like velcro.
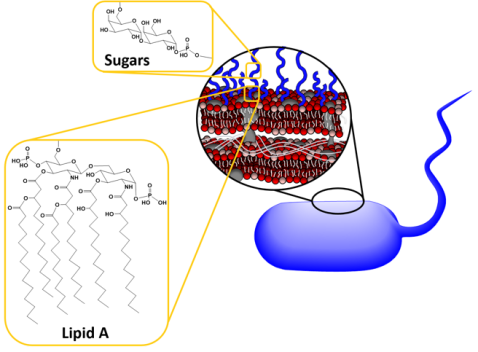
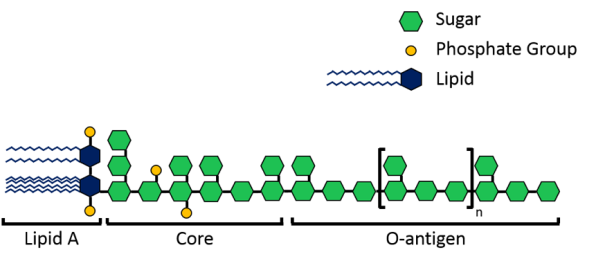
The velcro-like LPS molecules cover a large portion of the bacterial surface, and are important to the bacteria since they stabilize its structure and protect it from harmful chemicals. As shown in Figure 3 above, LPS molecules are made up of a lipid A region that anchors them into the membrane and then sugars that extend into the surrounding area. Now, each species of Gram-negative bacteria uses different sugars in its LPS, meaning that every species has a unique LPS structure. To add to that variation, bacteria can also have different lengths of LPS. Shorter molecules called rough LPS have a lipid A region and a short core sugar region, but longer molecules called smooth LPS have a lipid A region, short core sugar region, and also an O-antigen region, which is a long repeated sugar chain2 (Fig. 4).
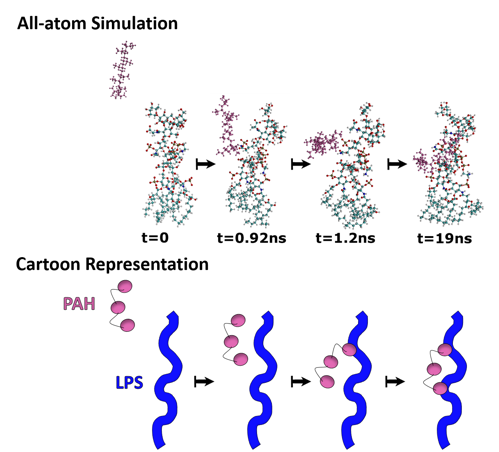
Since LPS are such a major player in the interaction of nanoparticles with Gram-negative bacteria, we wanted to learn more about the binding of PAH-wrapped gold nanoparticles with LPS molecules on bacteria. The way we did this was to perform computer simulations that take all the atoms in the whole nanoparticle-bacterial-membrane system into account, which is aptly named “all-atom simulations.” These work by taking the different properties for each unique atom into account in the calculation of how they will behave.
One limitation to all-atom simulations is how many atoms can be included, because the more that are included, the longer it will take a computer to do all the calculations necessary. If we tried to model a whole gold nanoparticle with a single bacterium, there would be over 83,000,000,000 atoms in our simulation!* Therefore, we created a simplified system where we looked at how the surface group on the gold nanoparticles, PAH, interacted with a single LPS molecule. This allows us to minimize the number of atoms in the simulation (to a little over 50,000), and yet each simulation still took about 65-75 hours to run!
What the first simulation showed us is that the PAH molecule (pink atoms in the image below) migrates to a particular part of the LPS structure: phosphate groups (brown atom surrounded by four red atoms). This told us that the phosphate is important for the PAH-LPS interaction (Fig. 5).
I mentioned before that nanoparticles bind to the bacterial surface and exhibit their toxic effects there, and so it’s easy to assume that more binding would correspond with more toxicity. The next step in our study was to move from computer modeling to a benchtop experiment to test the relationship between nanoparticle binding and toxicity. It was important to look at bacteria from different environments since nanoparticles can end up in many different environmental compartments (soil, freshwater, saltwater, etc.) and we wanted to see what effect nanoparticles would have in each of them. Therefore, we made a bacterial “panel” consisting of five bacteria from different environments that each have unique, beneficial environmental roles.
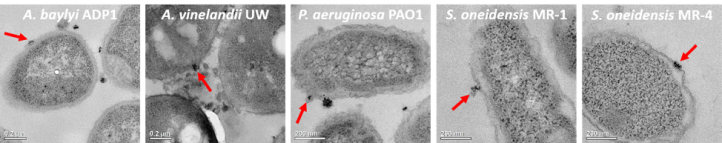
The first thing to do was to look at the nanoparticle binding to the bacteria, which is easier said than done because nanoparticles are too small to be visible with typical microscopes (for more on the limits of microscopy, see this blog post). We did this part of the study by exposing the bacteria to gold nanoparticles and then analyzing them with two methods. The first method was transmission electron microscopy (TEM), which allows us to visually see nanoparticle binding (Figure 6 shows the TEM images with a red arrow to indicate an example of nanoparticle binding for each bacterium in the panel). We also used a technique that told us how many bacteria had gold nanoparticles bound to them.
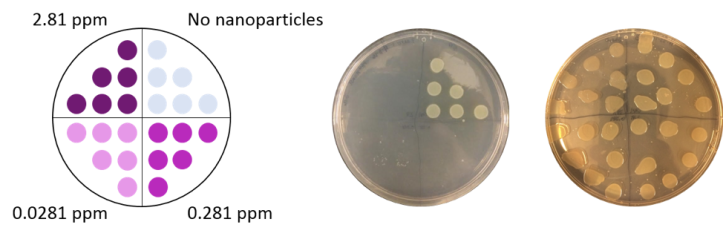
We also determined the toxicity of PAH-wrapped gold nanoparticles to each bacteria. This was done by determining what concentration of nanoparticles was required to kill the bacteria. To perform these studies, we exposed the bacteria to gold nanoparticles at a range of concentrations (0, 0.0281, 0.281, and 2.81 parts per million (ppm)). After the exposure we put six drops of each bacterial exposure on nutrient-rich plates for the surviving bacteria to grow. We then looked at which concentrations caused the bacteria to die. The bacterial growth appears as off-white/yellowish circles where the drop was placed, so if there are no circles we know the bacteria did not grow (Fig. 7). (This technique is similar to the “colony counting” technique I wrote about in this blog post.)
With the toxicity data collected, it was time to compare to the binding data we had. What we found in this comparison was not what we expected. Looking at our data, we saw instances where there was a similar amount of binding but very different toxicities, or where there were similar toxicities but different amounts of binding. Overall there was no correlation between the amount of nanoparticle binding and the toxicity observed.
The takeaway from this is that the more types of bacteria that are included in a study, the more we find biological complexity that belies the simple relationship of “more binding means more toxicity.” Identifying these biological complexities is important for understanding the full nature of the interaction between nanoparticles and bacteria. We don’t completely understand yet why more nanoparticle binding does not always mean more toxicity for bacteria, but our study demonstrates the importance of using a bacterial panel like ours instead of a single bacterial species for anyone who wants to do nanotoxicity studies in the future. Doing more studies to understand the complexity of nano-bio interaction should help scientists to predict what toxic effects nanoparticles might have on different organisms, which in turn will allow them to design nanoparticles that have reduced environmental impact.
FOOTNOTE
* For this estimate, I calculated the atoms in a bacterium to be 8.34×1010(using the mass of a bacterium3 and its elemental composition4) and I actually used our Sustainable Nano blog post about calculating how many gold atoms are in a gold nanoparticle (which, being around 2000 atoms, really doesn’t contribute to the overall amount of atoms in the bacteria-nanoparticle system; as the number of gold atoms was so small relative to the atoms in a bacterium, the atoms in the ligands on the nanoparticle surface were not calculated).
REFERENCES
- Joseph T. Buchman, Ali Rahnamoun, Kaitlin M. Landy, Xi Zhang, Ariane M. Vartanian, Lisa M. Jacob, Catherine Murphy, Rigoberto Hernandez and Christy Haynes. Using an environmentally-relevant panel of gram-negative bacteria to assess the toxicity of polyallylamine hydrochloride-wrapped gold nanoparticles, Environmental Science: Nano, 2018, 5 (2), 279-288. doi: 10.1039/C7EN00832E
- Wang, X.; Quinn, P.J. Endotoxins: Structure, Function, and Recognition. Springer: New York, NY. 2010.
- Davis, Dulbecco, Eisen, Ginsberg, Bacterial Physiology: Microbiology, Second Edition, Maryland: Harper and Row, 1973: 96-97.
- B10NUMB3R5, Empirical elemental formula for biomass (2017): http://bionumbers.hms.harvard.edu//bionumber.aspx?id=101800&ver=18

Great stuff, AND well-written!