What happens to cells when they come into contact with nanoparticles? Researchers in the Center for Sustainable Nanotechnology are trying to answer this question in a lot of different ways. The effect of exposing organisms to nanoparticles is not as simple as life or death. In one recent study1 we explored the molecular changes that can happen with an exposure level that is not enough to kill a living organism (otherwise known as a “sub-lethal” exposure). For example, organisms may respond to nanoparticles by changing the expression level of ribonucleic acids (RNA) in cells, and these changes affect cell and organism functions.
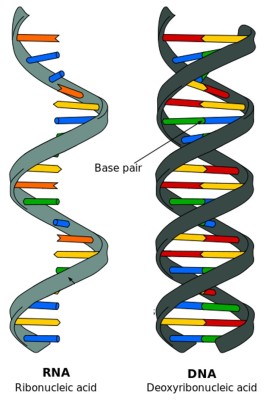
What do we mean by RNA expression level? You may remember that in living cells, deoxyribonucleic acid (DNA) carries most of the genetic information for the development, reproduction and function of an organism. DNA is double-stranded, as shown above in figure 1. It is the material that can replicate itself and can be passed on to the organism’s offspring. However, only having DNA itself is not enough; it’s like a manual with instructions of how a cell should function. By transcribing DNA into RNA, (shown in figure 2 below) a cell reads the manual and decides what actions should be taken. The cell adjusts the amount of different RNA molecules that need to be transcribed in response to different conditions, which is usually referred as the “expression level” of the RNA (also referred to as gene expression in some studies). After strands of RNA are produced, they are translated into proteins, and those proteins are the materials that finally execute cell functions.
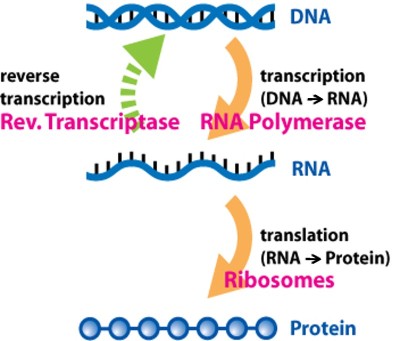
Because of this relationship between DNA, RNA, and proteins, it is natural for researchers to monitor the expression level of either RNA or proteins in order to figure out how cell functions change after they are exposed to changes in their environment (like the introduction of nanoparticles). In this study,1 we monitored how RNA expression (aka gene expression) changed in order to see how organisms responded to nanoparticle exposure. This technique has been used in past to discover what makes certain chemicals toxic. For example, researchers have found that pesticide exposure affects the expression of the gene for vitellogenin, an important protein in fish reproductive systems.2
In our recent project, we used a technique called quantitative polymerase chain reaction (qPCR) to study two specific types of organisms: the bacterium Shewanella oneidensis and a type of water flea called Daphnia magna (see figure 3). We extracted RNA materials from the bacteria and water fleas after exposing them to gold nanoparticles, and reverse transcribed the RNA into complementary DNA materials (as shown by the green dashed arrow in figure 2). Once we had the DNA, we used qPCR to determine the amount of genetic materials that was expressed. (The qPCR technique has long been used in a wide range of applications where identifying genetic expression is helpful, including diagnosing infectious diseases, detecting microbes for food safety and lots of laboratory research as well.)
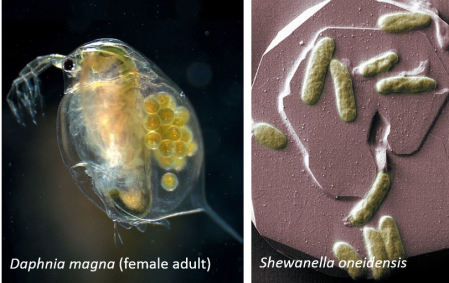
Monitoring the effects of sub-lethal dosages of toxic nanoparticles can give us a lot of useful information. The molecular responses we see using qPCR can be indicative of further damage to the organism that could affect reproduction and survival in the long run.
By learning about adverse effects like this, people can act to prevent them before it is too late. For example, we identified that the expression level of a particular RNA sequence called 16S in bacterial cells could be a potential indicator of reduced oxygen use in bacteria. This correlation still needs further testing, but if it is proved, we could use a decreased 16S expression level to predict that a certain nanoparticle would inhibit the oxygen uptake of bacteria at a higher exposure level. This could work even if the cells weren’t obviously inhibited or dying, and could help policy makers decide certain nanoparticles need to be regulated to prevent further accumulation before actual damage occurs.
Another great thing about using molecular/genetic responses to assess the risk of nanoparticle exposure is that you can examine these responses across multiple organisms. The biological diversity in our ecosystem can make it difficult to make general conclusions about how nanoparticles will affect a community of organisms, but a strength of our study is that we used two distinct model organisms (a multi-cellular water flea and a unicellular bacterium). We chose pairs of genes in each organism that encode for similar cellular functions, which allowed us to compare similar responses across organisms. When we exposed the organisms to positively-charged gold nanoparticles, we found that a stress-response system, called the heat shock protein response, was provoked in both of them. This might indicate that these nanoparticles affect different organisms in a similar way, at least in this one particular effect.
In addition to monitoring genetic expression in two different organisms, we also tested the effects of nanoparticles with the same core but different surfaces on each organism separately. We found signatures that could be a predictor of nanoparticle toxicity, and also features that were shared by both organisms. By understanding fundamental mechanisms of how organisms are impacted by nanoparticles, this research is building towards improving how we can assess and predict the outcomes of nanoparticle interactions with the environment.
This post is part of our ongoing series of public-friendly summaries describing research articles that have been published by members of the Center for Sustainable Nanotechnology. This collaborative study was done by researchers from University of Minnesota-Twin Cities and University of Wisconsin-Milwaukee. Tian (Autumn) Qiu, a doctoral student at the University of Minnesota, was a co-first author on this paper with Jared Bozich. The article was first published online in August 2015 in Environmental Science: Nano.1
EDUCATIONAL RESOURCES
- NOVA video: “What Darwin Never Knew: Switching Genes On and Off” (grades 8+)
- Concord Consortium DNA to Protein Lab: students can enter a DNA sequence & watch animation of its transcription to RNA and then translation to proteins. (Middle school +)
- Interactive Java simulation “Gene Expression – The Basics” from PhET.
- Ice Cream Sundae Gene Expression (grades 9-12)
REFERENCES (both available open access)
[1] T. A. Qiu, J. S. Bozich, S. E. Lohse, A. M. Vartanian, L. M. Jacob, B. M. Meyer, I. L. Gunsolus, N. J. Niemuth, C. J. Murphy, C. L. Haynes and R. D. Klaper. Gene expression as an indicator of the molecular response and toxicity in the bacterium Shewanella oneidensis and the water flea Daphnia magna exposed to functionalized gold nanoparticles. Environmental Science: Nano, 2015, advance article. doi: 10.1039/C5EN00037H.
[2] Nicolas, J. M. Vitellogenesis in fish and the effects of polycyclic aromatic hydrocarbon contaminants. Aquatic Toxicology, 1999, 45(2), 77-90. doi: 10.1016/S0166-445X(98)00095-2
