The awarding of the 2014 Nobel Prize in Chemistry to Dr.s Betzig, Hell, and Moerner (my former research mentor) is a tremendous event! It is almost as tremendous as their scientific targets are tiny: they were awarded the prize for super-resolution fluorescence microscopy, a technique for using a light microscope to examine objects as small as tens of nanometers. Dr. Christy Haynes talked about super-resolution microscopy in a recent blog post, so I won’t cover that again here. However, one of the enabling technologies for this year’s prize is the ability to use light to “see” individual molecules. Professor Moerner is a pioneer in what has come to be called “single molecule spectroscopy.” He is a true single-molecule zealot, and as a former trainee in his lab, I’m pretty much a single-molecule zealot as well. In this post, I hope to convince you that studying individual molecules is worth being zealous about.

First, we’ll talk about the “how.” The term “single molecule spectroscopy” might make it sound like we can use light to actually make out individual chemical bonds and atoms, but unfortunately that’s not the case.* Anyone who thinks that they are going to see a bunch of little “C”s and “H”s is going to be disappointed. What we can see is conspicuous, if indirect, evidence of the molecule interacting with light. In 1989, Professor Moerner published a paper about detecting individual molecules using a technique called absorption spectroscopy. In absorption spectroscopy, a whole bunch of photons are directed at the molecule, and a slightly smaller number of photons make it back out, minus the ones absorbed by the molecule under investigation. In a sense, Professor Moerner and his colleague Dr. Kador were able to see a “shadow” of a single molecule. They went on to publish some amazing work on how individual molecules can actually show different characteristics, even with the same molecular structure!
In 1990, Professor Michel Orrit published a paper using fluorescence to “see” individual molecules. The fluorescence used to see a molecule works kind of like a person standing in a dark room, pointing a bright flashlight right at your eyeballs. You know the person is there (because someone is holding that flashlight), but all you can make out is a bright blur! Similarly, the fluorescent molecule is emitting more photons than everything else around it, and the contrast is highly visible. In other words, you don’t see the single molecules themselves so much as you see the light emitted by single molecules. Being able to pin down the light’s location is an important principle for some methods of the Nobel-winning super-resolution microscopy.

But for single-molecule zealots such as myself, that’s not even the fun part. The flashlight in the analogy might not just stay steadily on. It could flicker on and off, or change color. The orientation (or polarization) of the light might be changing. Molecules go through all of these same maneuvers, and the details tell you fabulous things about how that molecule’s structure or environment is changing. You might wonder why it’s important to look at just one molecule for all of these fluorescent gymnastics. The answer comes back again to the person with the flashlight in the dark room. Imagine the flashlight is signaling Morse code. Now imagine a hundred other people in the room, all flashing different codes at the same time. Now you can’t pick out any individual message – the room isn’t even dark anymore! Molecules work the same way. All of the behavior we can detect through fluorescence is “unsychronized.” If you are viewing many (say, 10^23) molecules at one time, all you see is a boring and uninteresting average. But if you probe them one at a time, you can start to see all of their dynamic behavior. That’s what gets single-molecule zealots excited.

This technique can be used to learn a variety of different things about a molecule. For example, does a protein molecule have a specific group of atoms attached to it? If so, when does it attach, and how long does it stay? Another example would be using fluorescent color changes to detect when the distance between two molecules shifts, suggesting a change in the shape of the protein they’re attached to. My research group uses this phenomenon, called “Forster Resonance Energy Transfer,” to study the motions in proteins related to Alzheimer’s disease. Other groups examine a particular enzyme that seems to churn out products at random intervals. One of the beautiful parts of this work is that you don’t really know what the molecules will do next, which gives them a spooky and almost human quality.
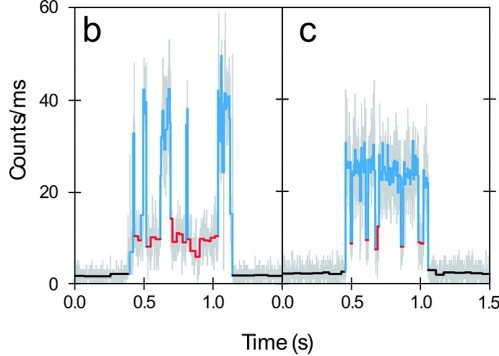
My drive to study single molecules comes from this ability to watch their dynamics occurring in real time. I gained much of my enthusiasm while training with Professor Moerner at Stanford University before starting my current position at the University of Wisconsin. Professor Moerner is an outstanding scientist and a wonderful human being. Hopefully some of those qualities have rubbed off on me as well. Congratulations W.E.!
NOTES
[*] There are ways to do this involving a beautiful class of techniques called “scanning probe microscopy,” although it is hard to apply them to see molecules moving or in a biological setting.
REFERENCES & FURTHER READING (subscription may be required)
Lee, Hsiao-lu D., Samuel J. Lord, Shigeki Iwanaga, Ke Zhan, Hexin Xie, Jarrod C. Williams, Hui Wang et al. “Superresolution imaging of targeted proteins in fixed and living cells using photoactivatable organic fluorophores.” Journal of the American Chemical Society 132, no. 43 (2010): 15099-15101. DOI: 10.1021/ja1044192
Moerner, W. E., and L. Kador. “Optical detection and spectroscopy of single molecules in a solid.” Physical Review Letters 62, no. 21 (1989): 2535. DOI: 10.1103/PhysRevLett.62.2535
Orrit, M., and J. Bernard. “Single pentacene molecules detected by fluorescence excitation in a p-terphenyl crystal.” Physical Review Letters 65, no. 21 (1990): 2716. DOI: 10.1103/PhysRevLett.65.2716
Wang, Quan, Randall H. Goldsmith, Yan Jiang, Samuel D. Bockenhauer, and W. E. Moerner. “Probing single biomolecules in solution using the anti-Brownian electrokinetic (ABEL) trap.” Accounts of chemical research 45, no. 11 (2012): 1955-1964. DOI: 10.1021/ar200304t

[…] As a life-long nerd and science-lover, it is hard to imagine a laboratory that could get me more excited than Galya Orr’s lab at the Pacific Northwest National Laboratory (PNNL). Galya is a collaborator within the Center for Sustainable Nanotechnology. I had the great opportunity to travel to her lab at the Environmental Molecular Sciences Laboratory (EMSL), which is a scientific national user facility located at PNNL, for two separate research trips in 2014. The trip to Richland, WA is worthwhile because Galya’s is no ordinary lab: it contains a veritable treasure chest of fluorescence microscopes capable of performing super-resolution techniques such as STORM, SIM (see a SIM tutorial here), and single molecule tracking, which are some of the same techniques that were awarded the 2014 Nobel Prize in Chemistry. […]